All Data Humanity Creates In A Year Stored On 4 Grams Of DNA
August 22, 2012
on
on

Data storage is becoming increasingly important as digital information doubles in volume roughly every two years. By the end of 2012 the volume will have grown 48% compared to 2011, the International Data Corporation (IDC) predicts.
Bioengineers have been jealously eyeballing nature’s information storage medium DNA for its efficiency and robustness.
Information stored in DNA can survive for a hundreds of thousand of years. Unlike data centers, DNA doesn’t need climate-control because it can withstand just about any environmental circumstance. As long as the data isn’t accessed there is no energy cost. And above all it has an extremely high storage density.
Now Harvard researchers report a major breakthrough. They have successfully encoded the contents of a book in DNA, copied it 70 billion times and fitted it on a space the size of a thumbnail.
For their experiment geneticist George Church and bioengineer Sriram Kosuri of the Wyss Institute for Biomedical Engineering at Harvard University used the book Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves written by Church. The HTML-version of the book, including 53,426 words, 11 images, and a JavaScript program was converted binary code resulting in 5.27 megabits.
The four types of bases of DNA adenine (A), cytosine (C), guanine (G), and thymine (T) each represent a binary value (A,C = 0, T,G = 1). Each binary value is represented by two bases to avoid bases of the same type being repeated more than three times.
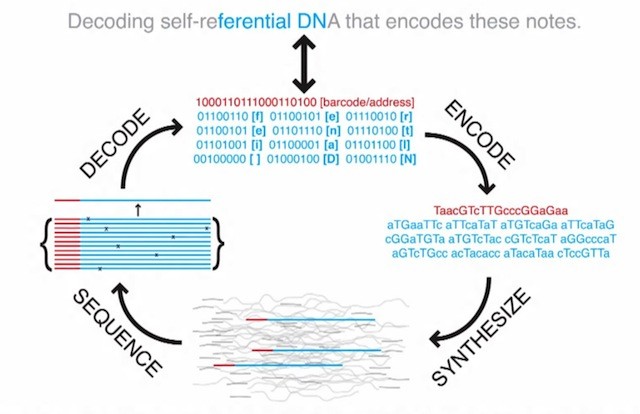
In the image you see a sentence from the HTML encoded book. A 12-byte portion of the sentence is converted into a data block of 96 bits (blue) preceded by a 19-bit address block (red). The bit sequence is encoded into bases, each base representing one bit.
The bases are then synthesized into strands of DNA on a DNA microchip.
To retrieve the data the DNA is sequenced (determining the order of the bases in a strand) and decoded to binary values. The address blocks are used to put the 96-bit data blocks in the right order.
The team stored the data on a synthetic DNA microchip. "We purposefully avoided living cells," Church said in the Harvard press release. "In an organism, your message is a tiny fraction of the whole cell, so there's a lot of wasted space. But more importantly, almost as soon as a DNA goes into a cell, if that DNA doesn't earn its keep, if it isn't evolutionarily advantageous, the cell will start mutating it, and eventually the cell will completely delete it."
DNA synthesizing and sequencing are slow processes therefore this form of data storage isn’t suitable for fast information retrieval. But it can be used for long-term storage and archival purposes.
The total volume of digital information in the world in 2011 was 1.8 zettabytes in 2011 (roughly 2 billion terabytes), IDC reported. As Kosuri points out in the video, all our accumulated knowledge could fit on 4 grams of DNA. It can then be tossed in a corner and left their for a couple of thousand years for our descendants to find and browse our collective knowledge. And if information keeps up its current growth rate they will marvel at how preciously little we knew.
Images: Clay tablet Hurrian Hymn Six. Source: Clingtgoss.com
Decode encode. Source: Supplementary Materials Next-generation Digital Information Storage in DNA. [The actual paper is behind a paywall].
Bioengineers have been jealously eyeballing nature’s information storage medium DNA for its efficiency and robustness.
Information stored in DNA can survive for a hundreds of thousand of years. Unlike data centers, DNA doesn’t need climate-control because it can withstand just about any environmental circumstance. As long as the data isn’t accessed there is no energy cost. And above all it has an extremely high storage density.
Now Harvard researchers report a major breakthrough. They have successfully encoded the contents of a book in DNA, copied it 70 billion times and fitted it on a space the size of a thumbnail.
For their experiment geneticist George Church and bioengineer Sriram Kosuri of the Wyss Institute for Biomedical Engineering at Harvard University used the book Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves written by Church. The HTML-version of the book, including 53,426 words, 11 images, and a JavaScript program was converted binary code resulting in 5.27 megabits.
The four types of bases of DNA adenine (A), cytosine (C), guanine (G), and thymine (T) each represent a binary value (A,C = 0, T,G = 1). Each binary value is represented by two bases to avoid bases of the same type being repeated more than three times.

In the image you see a sentence from the HTML encoded book. A 12-byte portion of the sentence is converted into a data block of 96 bits (blue) preceded by a 19-bit address block (red). The bit sequence is encoded into bases, each base representing one bit.
The bases are then synthesized into strands of DNA on a DNA microchip.
To retrieve the data the DNA is sequenced (determining the order of the bases in a strand) and decoded to binary values. The address blocks are used to put the 96-bit data blocks in the right order.
The team stored the data on a synthetic DNA microchip. "We purposefully avoided living cells," Church said in the Harvard press release. "In an organism, your message is a tiny fraction of the whole cell, so there's a lot of wasted space. But more importantly, almost as soon as a DNA goes into a cell, if that DNA doesn't earn its keep, if it isn't evolutionarily advantageous, the cell will start mutating it, and eventually the cell will completely delete it."
DNA synthesizing and sequencing are slow processes therefore this form of data storage isn’t suitable for fast information retrieval. But it can be used for long-term storage and archival purposes.
The total volume of digital information in the world in 2011 was 1.8 zettabytes in 2011 (roughly 2 billion terabytes), IDC reported. As Kosuri points out in the video, all our accumulated knowledge could fit on 4 grams of DNA. It can then be tossed in a corner and left their for a couple of thousand years for our descendants to find and browse our collective knowledge. And if information keeps up its current growth rate they will marvel at how preciously little we knew.
Images: Clay tablet Hurrian Hymn Six. Source: Clingtgoss.com
Decode encode. Source: Supplementary Materials Next-generation Digital Information Storage in DNA. [The actual paper is behind a paywall].
Read full article
Hide full article


Discussion (0 comments)